An interesting presentation by Ted Pedersen on finding obesity morbidities in i2b2 records.
Tuesday, December 29, 2009
Monday, December 14, 2009
German i2b2 implementations move forward
Thanks to Sebastian Mate for this update on the first German Academic i2b2-Workshop.
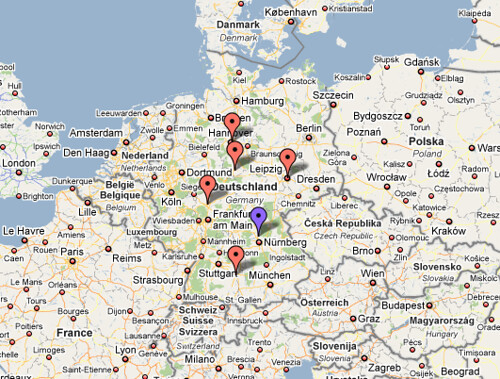
They had visitors from the universities of Goettingen, Ulm, Hannover, Giessen and Leipzig. Some of them had to travel hundreds of miles (see attached map) to visit them in Erlangen. Goettingen and Leipzig have i2b2 instances running, the others will try it out.
(1) Clinical Data Warehouse in Erlangen(2) i2b2 Overview, motivation and work in Erlangen(3) i2b2 in 30 Minutes - how the Erlangen Package works, with live-installation ;-)(4) The simple SQL-based Erlangen ETL approach - how we load data into the hive(5) i2b2 HIS-mapping - how we map hundreds of attributes from our HIS into the i2b2 instance (ONT and CRC)(6) TMF PID-Generator and i2b2(7) TMF Pseudonymization Service
Friday, December 11, 2009
RA
Discussed the SNP's on the genotyped data and how the continent of origin and the PHS labels of ethnicity are highly concordant.
Tuesday, November 17, 2009
Open Source
A very insightful response from Fred Trotter. He lays down the options in a very nuanced and clear fashion. Much appreciated.
Thursday, November 12, 2009
Major Depressive Disorder
Perlis, Smoller, et al.,
MInutes courtesy Patience Gallagher.
· Longitudinal Classifier
o Roy and Victor will finalize parameters of algorithm
o Victor will then report (and provide a visualization of):
§ % depressed, % well, # of all notes, etc
o To query Crimson: Victor will provide a list of medical record numbers within the two groups of interest (responsive and resistant) to Lynn Bry who can then report how many samples are currently available within Crimson
§ Parameters may need to be readjusted, depending on response from Crimson
· Discussion of Validation
o The issue with last week’s approach:
§ The algorithm is based on the text, so the first level of validation should not use information (i.e. clinician’s extensive knowledge of a patient) that is not in the text.
o New validation plan:
§ STEP ONE:
· GOAL: Determine if an expert clinician’s classification (based on notes only) is the same as the result of the algorithm
· Pull successfully classified notes
o All of a patient’s notes will be reviewed
o Clinicians will be blinded to the results of the classifier
o The sample of notes will reflect the results of the algorithm: “Random but representational”
o To keep the validation clean, it will be a “case-control” model - Treatment resistant vs. Responsive
o For now, only the electronic medical record will be used.
§ The consensus was that patient charts would be more annoying than beneficial, and the electronic records probably have sufficient information
§ Additionally, the information from the paper records is not integrated into the algorithm
§ May look at paper records down the road
§ STEP TWO: Compare list of patients that Roy knows are treatment resistant or responsive and run their notes through the classifier
· (Tianxi says this will be beneficial as it will give more power to the classifier)
§ STEP THREE: Use the Quick Inventory of Depression Severity (QIDS) as an external source of validation
· Many patients have QIDS scores in their charts – can determine if the algorithm classification is consistent with performance on the QIDS – this would be a cross-sectional measure
o The output of this approach would be: “Among patients classified s depressed, the mean QIDS score is ____”
§ NEXT STEPS:
· Do first level of the validation over the next few weeks.
o Victor will pass the notes to Roy.
o Roy will be the sole clinician reviewing the notes
· Manuscript:
o Victor and Tianxi have provided their input to Roy
o Roy will integrate this information and then re-distribute the manuscript to the group
· PV
o Update from Victor on obesity:
§ Compared the BMIs of this data set with all other patients and the distribution of the MDD sample is very similar, but right shifted compared to majority’s BMI distribution
· This makes sense! - Being depressed (and on antidepressants) leads to weight gain
Friday, October 30, 2009
Widening the Use of Electronic Health Records Data for Research
A symposium hosted by the National Center for Research Resources (NCRR). Louise Ramm, Deputy Director of NCRR provided framing challenges and welcome. Zak Kohane introduced use cases, sources of and reviewed the false dichotomy between health-record based research and clinical trials..
Gary Gibbons provided a perspective of disease in the African-American population as an exemplar of a complex orphan disease in the sense that like rare orphan diseases, it is understudied and insufficiently treated. He also pointed out how in many parts of the country underserved minorities are located away from the academic health centers that have made the most inroads in the use of electronic health records.. Therefore, institutions such as Morehouse School of Medicine have their work cut out for them (and not a lot of resources) to integrate data from a large number of only lightly affiliated practices. That same challenge presents an opportunity to be even more impactful in an orphan disease of epidemic quality. Professor Gibbons also urged a broadening of the captured context beyond what is conventionally captured in a standard (brief) healthcare visit. Environmental variables that are highly penetrant, much more so than many genomic markers are poorly captures. He concluded by reviewing the current compelling information about pharmacogenomic differences, and population genetic risks and also the wide holes in our knowledge of these as they pertain to various groups within the USA.
Andrew Auerbach from UCSF addressed comparative effectiness research and its translation into "Health system innovation research" Described how much can be done with charge data, and how additional codified data types (e.g. medications) can further improve the quality of that data. Closed with a discussion of how the various stakeholders in using EHR data for research (.e.g NIH, Payors, health systems leaders, physicians, and patients) might be well aligned or not. Put us on notice that IRB's are unfamiliar about distributed query systems and/or grids and this is becoming at last an obstacle for many CER studies. Summarized several use cases such as optimal length of treatment of pneumonia? Can a patient-focused discharge checklist reduce risk for readmission?

Robert Plenge described his use of i2b2 and electronic health records for genotypic research and discovery of endophenotypes.
John Brownstein reviewed non-traditional public health research using institutional data and non-traditional, non-institutional healthcare data extraction and analysis.
Wednesday, October 28, 2009
i2b2 Academics Users Group—Natcher Building, NIH
Zak Kohane summarized some of the new i2b2-based projects that recently were announced including a South Carolina consortium (a GO grant funds the automated consent component), a pediatric rheumatology research network including 60 sites (NIH GO grant), Shawn Murphy is working with an imaging consortium to better integrate images into i2b2 instances (CTSA administrative supplement) using XNAT/BIRN infrastructure, and B.U. and U. Mass received a GO grant to study health disparities using i2b2 as infrastructure. Lynn Bry received a 2 year R01 under ARRA for the Crimson-i2b2 integration project.
Susanne Churchill welcomed the group and noted that there is a consortium of European i2b2 users/implementors/refiners that are putting together a proposal for joint work across the European Union (for EU funding). She noted that the AUG now numbers over 100 and represents 30 healthcare/academic institutions including 5 internationally.
Shawn Murphy summarized several new developments including
- Release Candidate 1.4 to support the Enterprise including: analysis views, improved role-based access and auditing, replacement of Gridsphere with webservices and AJAX client, Microsoft Active Directory integration, obfuscation of results for privacy purposes.
- In the future, the distributions of i2b2 be via configurable VM's so that the functionality can be attached to local databases without requiring a full install. The full source code compile and install will still be available.
- Eclipse plug-in "store" where developers can contribute their own plug ins and where users can download the plugins they want.
- More support for derived data (e.g. to systematically return NLP concepts derived from the clinical notes
Lynn Bry described the Crimson system and how i2b2 discarded sample have far higher utilization rates than other samples (e.g. for biorepositories). She also described the Sample Ontology and how she is borrowing from WHO and SNOMED to standardization. The system includes an ontology manager to allow local ontology management and update samples. Also described are the IRB permissions data from RC 1.4. Will work towards multisite studies within the two year implementation time frame. Lynn described the Enterprise Master Specimen Index that tracks samples and patient relationships in various levels of identity (consented identified, de-identified, and anonymous samples).
Dan Housman and Peter Emerson from Recombinant were invited into the AUG for the discussion segment about sample management (because of the AUG's wish to keep companies at arm length) to discuss their own efforts in sample management. They made it clear that all their developments they are involved in will be contributed back to the i2b2 community as fully open source code.
Andy McMurry summarized the status of the distributed querying system called SHRINE that is now implemented at several Harvard-affiliated hospitals and several West Coast academic health centers (e.g. UCSF and UW) that is now fully IRB approved (at Harvard) for queries returning aggregate numbers (across demographics, laboratory results, medications and diagnoses). Andy also made a very clear several technical hurdles that were overcome including the ontology matching process (on the fly). Finally, he announced the availability of SHRINE code in a fully open source codebase.
i2b2 CICTR presented by Nick Anderson. They have been able to query muiti-institutional "anonymized PHI data". Application is in diabetes and cardiovascular disease. Described technical, governance, ontology and evaluation process that CICTR is driving. Nick described the heterogeneous systems that CICTR has to query across. Nick distinguished the need for high level institutional support which is a sine qua non requirement for success and the need for a broad range of paid technical personnel.
Keith Marsolo from Cincinnati's Children's reported on their Epic roll out and how that relates to their i2b2. Described their quality assurance efforts. Notes the challenge of the firehose and makes the acute observation that most investigators just want a spreadsheet and anything more complicated than that tends to get ignored. Keith also emphasized their goal to allow streamlined adding of research data to the clinical data. He makes the important point that "age at FACT" is essential for pediatric applications to allow them to be easily accessed in the i2b2 workbench. Keith mentioned using i2b2 for research databases for Eosinophilic esophagitis, and IBD.
Phil Reeder from UT Houston talked about medications mapping. It is a challenge and they have chosen to map to RxNorm and then manually had to map into SNOMED CT (perhaps their database was out of date). Started from an All Scripts database and had a semi-automated process. Notes that every year there are at least a 1000 new drugs (i.e. different packaging, pill sizes etc). Brought up the thorny (and annoying IMHO) of the proprietary mappings to standard vocabularies.Ralph Zottola and Edward Westrick described the effort at U. Mass (data sourced from Meditech system, REDcap EDC, biorepository, EMPI, Allscripts, and departmental systems) where they are up to 2,000,000 patients. Edward described the managed care network (1000 physicians) that plugs into U. Mass and how quality measures inform the discussions and bargaining with payors. Reviewed different measures including HEDIS, patient experience, was well as the increasingly important Relative Resource Utilization (Efficiency). Demonstrated how knowing what is going on in the healthcare institution allows for a sober and leveraged discussion with payors. The healthcare system approached the medical school and settled on i2b2 and they already have seen that they can accurately forecast their performance and to provide a feedback loop (with financial incentives) to healthcare providers. Ralph pointed out that the fact that clinical operations are using i2b2 is also causing an improvement in the quality of the data being delivered to the data marts.
Iain Sanderson and Jihad Obeid. Iain started by describing a very comprehensive Informatics Initiatives in South Carolina. They have a unified IRB with a goal of clinical trials across the state. There is both a scientific and a funding motivation in this. There are three informatics initiatives have dovetailed (CTSA biomedical informatics, HSSC IT business plan, and a GO grant on consent). This has resulted in the South Carolina Integrated Platform for Research (SCIPR) that uses i2b2 for the clinical research data warehouse. In the process they are adopting a wide range of open source solutions including Sun Microsystems' JavaCaps. Iain reports that the data sharing agreements between the 6 centers across HSSC are under way and likely to result in an MoU in short order. Iain also described the beginnings of the consent management/gathering system, the permissions ontology and documenting the different consent processes at the institutional members of the HSSC. Finally, Iain discussed how personal patient health portals may be used to provide the patient-facing part of the network.




Friday, October 23, 2009
Rheumatoid Arthritis
Plenge et al.,
3/4 of the genotyping is completed.
Manuscript to be submitted today regarding phenotyping accuracies.
What's next
Discussed what might be the priority areas for i2b2 in Core 2 for the competitive re competition.
The RFA has not yet be announced so the discussion was, of necessity, wide-ranging.
Major Depressive Disorder (and Bipolar Disease)
Smoller, Perlis, et al.,
Discussed the challenge of merging note-level NLP conclusions to wholistic patient evaluations.
Reviewed various Bayesian and (more broadly) machine learning approaches to defining which notes contribute most to the accurate classification of the patient phenotype.
Major Depressive Disorder: 10/16/09
Courtesy Patience Gallagher
Minutes:
Collapsing terms
o McLean admission à change to psych admission
o List of terms that Jordan gave Margarita that we didn’t necessarily annotate, Margarita will add after.
o Terms that have the same regular expression will be filtered at the end
o Medication category: fga
- We annotated as IW bipolar, but last time we said was CW, which one is it?
- Sergey says right now doesn’t matter, because the computer will decide
- We will keep it labeled as IW
o Li/vpa/lamtical à change to mood stablilizer
- Add term mood stabilizer(s)
o Neuro/cognitive impairment includes
- Confused and disoriented
- Gross cognitive impairment
- Significant cognitive deficits
- dementia
o Agitation includes
- Hyperactivity
- Hyper
- pacing
o dx depression includes:
- depressive disorder
- dsythymia
o bipolar disorder includes:
- dx bipolar disorder
- 296.0, .1, .4, .5, .6, .7
-
- .2, .3 are MDD, so we cannot say 296.x = BPD
o Inappropriate behavior category, leave in:
- Wanting to disrobe
- Inappropriate sexual contacts (do not put with excessive pleasurable activities)
o Rapid cycling
- 4 or more per year
o Mania will include
- Category now called “mania/manic”
- Category now called “manic episode”
- Cycling, cycles (not specified as rapid/less than 4 per year)
o Typos will include variations on spellings of:
- Grandiose
- distractibility
o “loose associations” will be its own category
o Depressed episode à put with hx depression
o “unable to read, study, concentrate” à put with distractibility
o “tx bipolar disorder” à add in term “rx”
o Hypomania will be own category
o hyper mood à mood elevation
o Delete:
§ Line 310: dx substance abuse, schizophreniaParanoid schizo-affected disorder
§ Out of control
Big group: 9:30-10:30
· Protocol in the grant
- Are we doing what we said in the grant? For ex. Step #3
-
- In grant, describes that classic way of doing NLP
- You come up with list of terms from your head without looking at notes.
- Then expand these terms, add regular expressions, negation
- Our method
-
- Is a hybrid of the 2. We annotated notes, but we also came up with a list of terms
- Annotate notes, generate list of terms, create regular expression, group, add in a priori terms, Tianxi goes through them and takes out terms that don’t matter (lasso), then feed to the computer
- The IRB will not really care that this is slightly different.
- Which method is better?
-
- Unknown
- Classic model relies on what’s in your head, our method relies on # notes reviewed
- We could try to compare, but it’s a little too late because our list of terms wasn’t blind to the notes
- Dr. Savova ) will be coming on boad as NLP team lead
-
- Would be good for her to give mini lecture about NLP so we are prepared to present at conferences
- Should consult with her about project
- Negation
-
- So far Margarita has only added negation to some terms
- Should modify algorithm so that every term also has corresponding negation term
- Negation terms are very common in psych notes
- Sergey to see if this can be done, although a lot of work
- Would be useful for future i2b2 projects
- Margarita explained that she plans on conducting a small validation study
-
- For a particular patient, she can provide an idea of how strong the diagnosis we assign (e.g. BP2) is by showing what are the arguments for/against placing a person in a particular class
· Next steps:
o Jordan, Vivian, Margarita, Sergey, Victor, Roy to meet on Tuesday, October 20th 10:00 – 11:00 to finish collapsing term.
Friday, October 9, 2009
Rheumatoid Arthritis
Plenge et al,
384 genotypes of risk alleles from prior studies.
A subset of the genes will shortly also have their exons resequenced.
Discussed adding antibody studies to the RA datamart. As a preliminary to adding the SNP and other data sets.
Major Depressive Disorder
Perlis, Smoller, Iosifescu et al.,
Discussed the new privacy guidelines that come with the new Recovery Act. This may make it harder for those of us who are trying to advance medical science and it is unclear if it will slow down those who are trying to commercially exploit patient data. What it is certain is that it will increase the workload/opportunities for lawyers specializing in medical privacy.
Victor reported that he is running Tianxi's logistic regression model of the NLP features on the 2M notes in the MDD data mart.
Discussed the cross-DBP NLP challenges worth investing some hardening on. So far this includes cigarette smoking, alcoholism, obesity, and other substance abuses.
Open source
Discussed planning for going from the current open source license to open source community support.
Friday, October 2, 2009
MDD
Perlis, Iosifescu, Smoller et al,
Reviewed claims data vs (claims data + NLP) and saw very large increase in AUC for treatment response.
Also discussed recruitment rates with much tighter inclusion criteria.
i2b2 AUG prep
Shawn, Diane, Zak, Susanne, Griffin
Discussed provisioning new DBP's
Discussed the GO grants that other sites have obtained using i2b2 infrastructure.
Friday, September 25, 2009
Rheumatoid Arthritis
Kat Liao et al.,
Reviewed our edits to the manuscript that Kat is pulling together. Discussed the underlying hypothesis.
MDD
Smoller, Perlis, Iosifescu et al.,
Tianxi showed that the incremental value of billing codes to NLP'd characterizations of depressed patients was minimal at a wide range of false positive rates.
Then a longer discussion regarding controls.
- Utilization is a bad match, because increased utlization is confounded with psychiatric overlay.
- Completely healthy individuals is a bad match because we would be selecting for populations with a lower burden of risk alleles for numerous diseases.
(The following courtesy of Patience Gallagher):
Isaac Kohane, Susanne Churchill, Jordan Smoller, Roy Perlis, Sergey Goryachev, Shawn Murphy, Dan Iosifescu, Victor Castro, Tianxi Cai, Vivian Gainer, Wouter Hoogenboom, Margarita Sordo, Stefanie Block, Patience Gallagher
Monday, September 21, 2009
Friday, September 18, 2009
Short term planning
Zak, Shawn, Griffin, Susanne
Committed to the details of AUG meeting at the CTSA
Reviewed additional hires required for ancillary i2b2
Discussed storage needs.
Wednesday, September 16, 2009
MDD
Minutes courtesy Patience Gallagher
I2B2
Meeting Minutes
Date: Friday, September 11, 2009
Attendees:
|
Vivian Gainer |
Sergey Goryachev |
Dan Iosifescu |
|
Shawn Murphy |
Roy Perlis |
Holly Sciortino |
|
Jordan Smoller |
Margarita Sordo |
A
Minutes:
- Scheduling a reoccurring meeting to discuss the RO1s (ICCBD & Roy’s RO1)
-
- Roy and Jordan will forward suggested days/times to Susanne Churchill to decide on
- Imaging (Dan):
-
- Update on analyses:
-
- Good data from the structural images
- Continuing to work on DTI
- Dan is still waiting on healthy volunteer data from Vivian
-
- Vivian reported that the delay has been due to the source system changing the format of the RPDR data. However, since the team only wants the reports and not the images, Vivian anticipates being able to provide the data to Sergey by Monday (September 14, 2009).
- Bipolar
-
- The best model is the B0, B1, B2 vs. NB2
-
- Exact features used include: bipolar disorder, Depakote, grandiose thinking, lithium, major depressive disorder, irritability
- Grandiose thinking appears to predict a non-bipolar diagnoses
- Predictors
-
- Each term highlighted by the diagnosticians is being treated as a separate predictor
-
- For example: Although “Family History: Bipolar disorder” and “Family history of bipolar disorder” mean the same thing, they are being counted/grouped separately.
- Margarita will review the terms and group the expressions which will increase the frequency of the predictors
- Improving the performance of the model
-
- Layering additional requirements on top of the model would increase the features used to determine the diagnosis
-
- For example: Require that in order to be labeled as Bipolar, a prescription of “Lithium” would need to be found in at least 3 separate notes
- Jordan will provide additional features to layer on top of the model
- Interpreting the model (B0, B1, B2 vs. NB2)
-
- Why isn’t B2 vs. NB2 the best model?
-
- Could be due to the number of instances
-
- 69 cases included in the B2 vs. NB2 model versus 84 cases in the NB2 vs. B0, B1, B2 model
- The current “best model” isn’t doing what we ultimately want
-
- The “best model” could change once Margarita groups the features
- If this doesn’t work, we may need to determine if additional notes need to be reviewed by the diagnosticians
- Predicting cases versus controls
-
- Will we be able to determine controls without an algorithm?
-
- Everybody not identified by the bipolar algorithm as a B2 would be considered a control
-
- This is not a good solution, since we want people that don’t have evidence of any psychiatric conditions
- If we only identified individuals that don’t have psychiatry notes, this would not necessarily mean that they don’t have a psychiatric condition rather we may be identifying individuals that don’t use the health system
- Shawn suggested matching controls by age and gender and then excluding certain diagnoses
- When reporting the algorithm’s performance at selecting Bipolar cases and distinguishing them from controls we should report the percentage of individuals with Bipolar disorder we would expect from the MGH population and compare that to the population prevalence
-
- Roy suggested identifying 200 individuals classified as controls by Shawn’s method (above) and using a bipolar screening instrument to determine if they are truly controls
-
- Unsure of how we would recruit these individuals
- If there are a few true cases included in the control sample, there would be a small effect. Controls that are identified as cases have a huge affect in research on diseases with low population prevalence (such as bipolar disorder)
- Things to look into:
-
- Margarita will look into why grandiose thinking is not predicting bipolar disorder
- Margarita/Sergey will reconfirm that the model is not just looking at diagnostic codes
- Margarita will group the features
- Margarita/Sergey will layer further features on top of model (after Jordan provides the additional features)
- Team will determine if we need to review more cases
- MDD
-
- How will we determine responder vs non-responder?
-
- Vivian ran analyses per subject based on billing codes and created patterns according to the timeline of the diagnoses, visits to psychiatry, and prescriptions
-
- Potential Issues:
-
- The diagnostic codes are not used consistently in the notes
- When the visits to psychiatry end, unsure if the patient is lost to follow-up, in remission, or seeing a private psychiatrist
- If ECT is present at the same time as a prescription, unsure of which treatment the patient is responding to
- Additional criteria may be needed such as death
- Potential benefits:
-
- Once above issues are addressed, we may be able to look at the patterns to determine responders vs non-responders
- It would be informative to add NLP to this visual
-
- Tianxi will take the last digit of the diagnostic code, extract the status and incorporate into the model/algorithm
- If the results are very different, we may have evidence that research based on billing codes is missing data and therefore inaccurate
- Categories/Parameters
-
- Roy and Dan drafted parameters which may be able to add complexity to the classifier
- Roy will send these to Shawn
NEXT MEETING: 9/18/09
Friday, September 11, 2009
Review of Core 1 needs from Core 2
Murphy et al.
Reviewed data marts required for Core 1 activities
- Normals
- NLP
- Predictive medicine
- Relevance networks
- Inflammation as an underlying process across diseases (the systems approach)
Friday, September 4, 2009
Rheumatoid arthritis
Plenge et al.,
How to go from GWAS to prediction.
Discussed how to get better insight into the genetic "dark matter" that might explain additional inherited variation compared to published variants.
Major Depressive Disorder
Perlis, Smoller et al.
Discussed the NLP again and reviewed Tianxi's analysis of the NLP PPV and NPV and AUC
Sergei and Jordan reviewed the results of logistic regression trained on several hundred expert review cases.
Working on Web Client for PM Cell
Present: Shawn, Griffin, Susanne, Diane, Zak, Mike
Discussed a bare-bones web client for PM management.
Monday, August 31, 2009
Major Depressive Disorder (8/7/2009)
Roy Perlis
Holly Sciortino
Patience Gallagher
Isaac Kohane
Susanne Churchill
Vivian Gainer
Griffin Weber
Dan Iosifescu
Shawn Murphy
Sergey Goryachev
Action Items:
|
Action Item |
Description |
Owner |
Source of Action |
|
1 |
Ask Lynn if it's possible to query Crimson to see which imaged subjects blood is available for |
Roy |
8.7.09 Meeting |
|
2 |
Generate list of useful scans |
Dan |
8.7.09 Meeting |
|
3 |
Results of classification (outcome measures) |
Tianxi |
8.7.09 Meeting |
|
4 |
Make list about what concepts/covariates are needed to pull out |
Dan and Roy |
8.7.09 Meeting |
|
5 |
Move classifier to another workstation |
Shawn/Sergey |
8.7.09 Meeting |
|
6 |
Eventually move classifier to I2B2 server |
Shawn/Sergey |
8.7.09 Meeting |
|
7 |
Provide instructions for how to get to classifier using remote desktop |
Shawn/Sergey |
8.7.09 Meeting |
|
8 |
Talk with Dan Mirel – confirm genotyping costs |
Roy |
8.7.09 Meeting |
Minutes:
· IMAGING
i. Non DTI: Healthy volunteers(?): 8 out of 80 usable
1. total of 20 subjects anticipated
2. ~ 1 month to complete phenotype data set
ii. DTI: 170 out of 320 usable
1. total of 150 subjects (excluding children) anticipated
2. ~6 months to complete phenotype data set
iii. 5-10% of these will not be useful (but information about cases not included will be kept to reduce bias)
· Access to DTI data
· Classification
i. Shi and graduate student are working on using notes to determine outcomes
ii. Roy and Dan need to create a list of concepts/covariates to pull out of notes
· A new analyst will be joining the team soon – working under Vivian – but all work still goes through Vivian (She is the point person for DBP and PV)
· Sergey’s classifier
i. According to PHS security guidelines, identified data can only be stored on a PHS server
1. Data can not be open to databases without required approvals
ii. Short term solution = the classifier is currently only on Sergey’s workstation which can be accessed with remote desktop from a Partners’ computer or with VPN access
1. Shawn and/or Sergey will also provide instructions for how to get to the classifier using remote desktop
iii. Long term solution – Shawn plans to integrate the classifier with the i2b2 applications
· PV
i. Not much dose-response, but this is without covariates
ii. Use of multiple antidepressants leads to better signal for most of the poor treatment outcomes
iii. Roy (and others?) need to sort out how to do analysis
1. define clinical terms
· Genetics
The genotyping is expected to take 6 months (start to finish)
October 1, 2009 – Target date to cut the link and move samples over (independent of having imaging data available)